EPJ B Highlight: A new method provides better insights into real-world network evolution
- Details
- Published on 28 August 2017
Chinese scientists show how the network structure affects the accuracy of methods predicting the future evolution of a network, like those used to predict protein interactions
Nature and society are full of so-called real-world complex systems, such as protein interactions. Theoretical models, called complex networks, describe them and consist of nodes representing any basic element of that network, and links describing interactions or reactions between two nodes. In the case of protein-interaction studies, reconstruction of complex networks is key as the data available is often inaccurate and our knowledge of the exact nature of these interactions is limited. For reconstruction of networks, link predict -- the likelihood of the existence of a link between two nodes -- matters. Now, Chinese scientists have looked at the influence of the network structure to shed some light on the robustness of the latest methods used to predict the behaviour of such complex networks. Jin-Xuan Yang and Xiao-Dong Zhang from Shanghai Jiao Tong University in China have just published their work in EPJ B, providing a good reference for the choice of a suitable algorithm for link prediction depending on the chosen network structure. In this paper, the authors use two parameters of networks—the common neighbours index and the so-called Gini coefficient index—to reveal the relation between the structure of a network and the accuracy of methods used to predict future links.
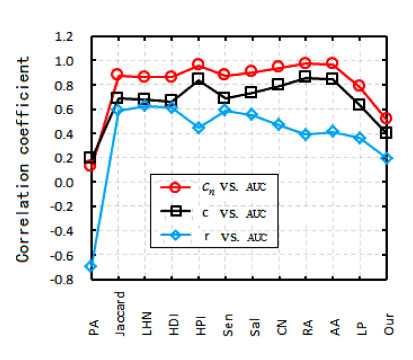
Their study partly involves a statistical analysis, which reveals a correlation between characteristics of the network, like the common neighbours index, Gini coefficient index and other indices that specifically describe the network structure, such as its clustering coefficient or its degree of heterogeneity.
The authors test their theory experimentally in a variety of real-world networks and find that the proposed algorithm yields better prediction accuracy and robustness to the network structure than existing methods. This also leads the authors to devise a new method to predict missing links.
Jin-Xuan Yang and Xiao-Dong Zhang (2017),
Revealing how network structure affects accuracy of link prediction,
European Physical Journal B, DOI: 10.1140/epjb/e2017-70599-4





