EPJ B Highlight - Fractal patterns in growing bacterial colonies
- Details
- Published on 18 September 2019
A new agent-based computer modelling technique has been applied to the growth and sliding movement of colonies of bacteria
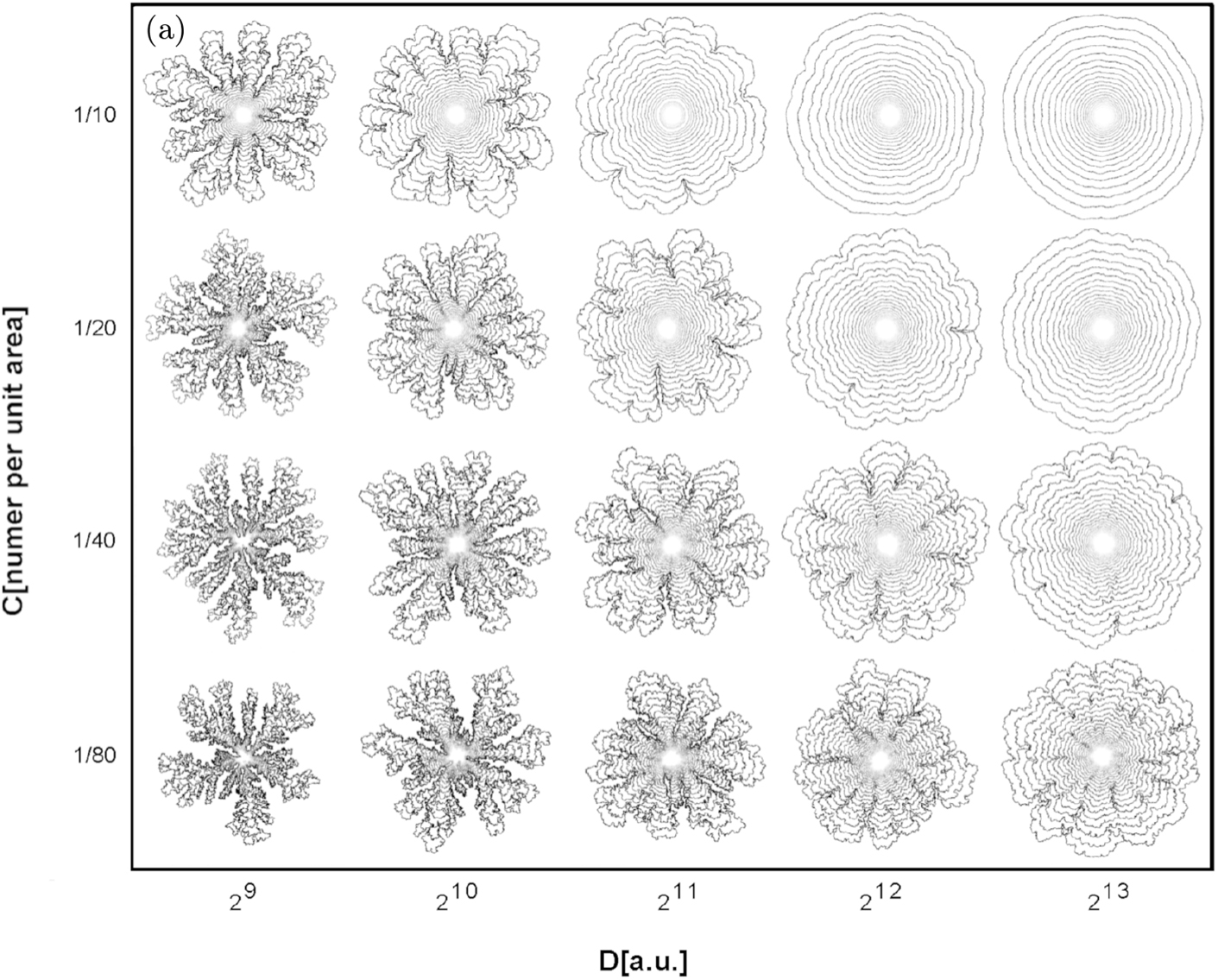
As many people will remember from school science classes, bacteria growing on solid surfaces form colonies that can be easily visible to the naked eye. Each of these is a complex biological system in its own right; colonies display collective behaviours that indicate a kind of 'social intelligence' and grow in fractal patterns that can resemble snowflakes. Despite this complexity, colony growth can be modelled using principles of basic physics. Lautaro Vassallo and his co-workers in Universidad Nacional de Mar del Plata, Argentina have modelled such growth using a novel method in which the behaviour of each of the bacteria is simulated separately. This work has now been published in the journal EPJ B.
A bacterial colony grows from a single cell, so all the bacteria are genetically similar clones of that original cell. Vassallo and his team simulated this pattern of growth on computers while varying different parameters: 'biological' ones such as the speed of cell division and the availability of nutrients, as well as 'physical' ones such as the mechanical forces between neighbouring cells. Their results agreed very well with patterns that have been observed experimentally. In the simulation, as in nature, all colonies began as compact round blobs with the snowflake-like fractal patterns emerging at a later stage.
The researchers used a multi-fractal analysis technique to describe the patterns produced by one specific type of bacterial movement: sliding. This means that the bacteria don't move independently but push each other within the colony by dividing and competing for the same space. This is only one of at least six well-defined types of bacterial motion, but it is particularly important as colonies use it to form persistent and medically challenging biofilms. Vassallo and his co-workers expect to apply their technique to simulating the other movement types, however, and even to modelling communication between bacteria within a colony.
L. Vassallo, D. Hansmann and L. A. Braunstein (2019), On the growth of non-motile bacteria colonies: An agent-based model for pattern formation, European Physical Journal B 92:216, DOI: 10.1140/epjb/e2019-100265-0




