EPJ E Highlight - Which sequences make DNA unwrap and breathe?
- Details
- Published on 04 December 2017
New study elucidates the DNA sequences that offer the perfect conditions for packaged DNA to unwrap and ‘breathe’, thus allowing genes to be read
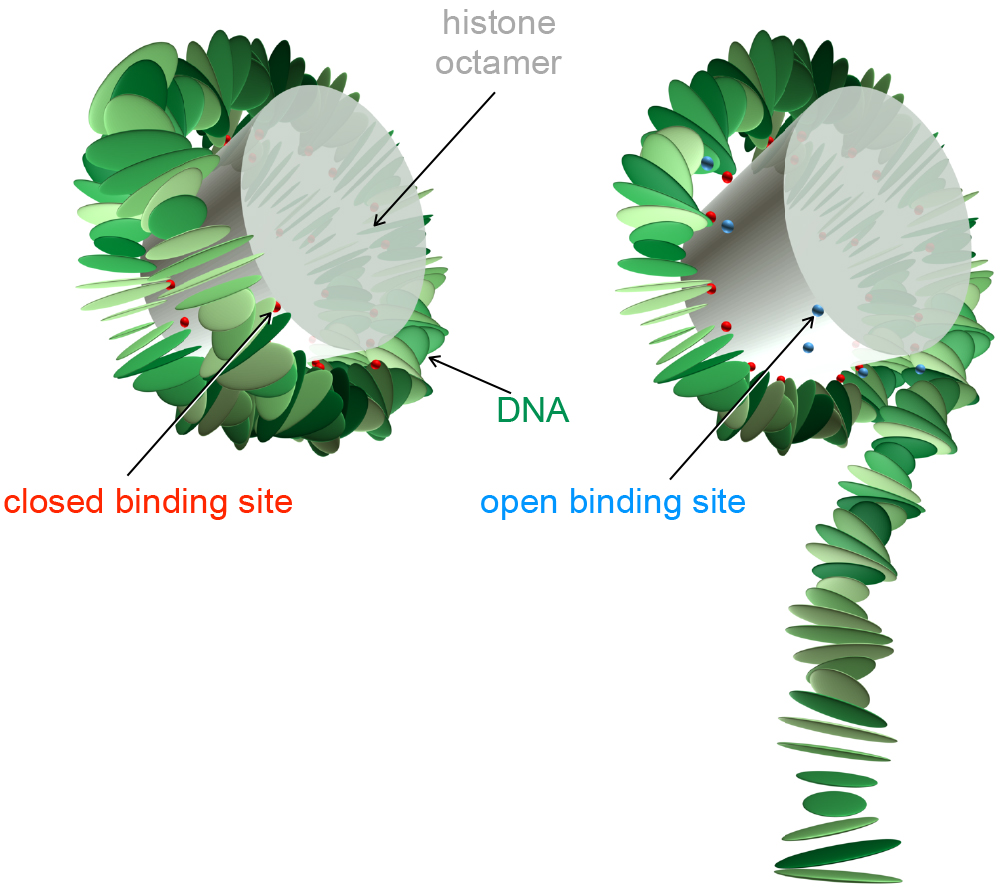
Accessing DNA wrapped into basic units of packaging, called nucleosomes, depends on the underlying sequence of DNA building blocks, or base pairs. Like Christmas presents, some nucleosomes are easier to unwrap than others. This is because what makes the double helix stiffer or softer, straight or bent—in other words, what determines its elasticity—is the actual base pair sequence. In a new study published in EPJ E, Jamie Culkin from Leiden University, the Netherlands, and colleagues demonstrate the role of the DNA sequence in making it possible for packaged DNA to open up and let genes be read and expressed.
Back in 1995, the late biochemist Jonathan Widom, from Northwestern University, Illinois, USA, demonstrated that nucleosomes ‘breathe’ for a short time, as the DNA partially unwraps from the protein cylinder. This is due to fluctuations linked to temperature changes. This past experiment measured quite accurately the relative accessibility of different parts of the DNA spool for enzymes that cut that DNA.
Following in his footsteps, the authors developed a model that uses computer simulations and is based on their own previously published coarse grained model of the nucleosome. The previous work posited the existence of a second layer of information in DNA molecules, one that is mechanical in nature. By contrast, in this study, using the nucleosome model with sequence-dependent DNA elasticity, the authors studied the effect of the base pair sequence on the accessibility of the nucleosome for a gene readout. Their approach yields a detailed explanation of how the underlying DNA nanomechanics dictate the physical properties of the nucleosome.
Incidentally, the model could also be used to interpret new studies related to the gene editing technology CRISPR, which is applied to DNA wrapped into nucleosomes, and relies on bacteria acting as an immune system that detects and destroys foreign DNA.
The role of DNA sequences in nucleosome breathing. J. Culkin, L. de Bruin, M. Tompitak, R. Phillips, and H. Schiessel (2017), Eur. Phys. J. E 40: 106, DOI 10.1140/epje/i2017-11596-2





